Data Related Infomation
Camellia sinensis var. sinensis cv. Shuchazao TEA006665.1 TRI CSS Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | No Gene Product Found |
| Locus Name: | TEA006665.1 |
Gene Family
ELFV_dehydrog,ELFV_dehydrog_N
Gene Attributes
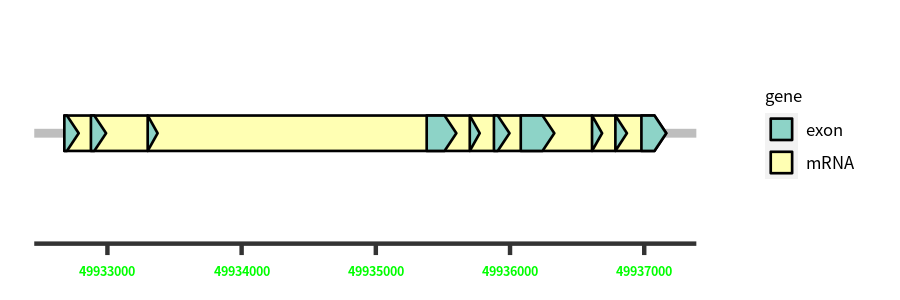
| Chromosome: | chr13 |
| Cds Coordinates: | 49932679-49937164 |
| Nucleotide length: | 4485 |
| Prodicted protein length: | 436 |
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| Pfam | Glutamate/Leucine/Phenylalanine/Valine dehydrogenase | 201 | 433 | GO:0006520|GO:0016491 |
| PIRSF | Glu_DH | 94 | 436 | GO:0016639 |
| PIRSF | Glu_DH | 1 | 98 | GO:0016639 |
| PRINTS | Glutamate/leucine/phenylalanine/valine dehydrogenase signature | 361 | 372 | GO:0006520|GO:0016491 |
| PRINTS | Glutamate/leucine/phenylalanine/valine dehydrogenase signature | 192 | 214 | GO:0006520|GO:0016491 |
| PRINTS | Glutamate/leucine/phenylalanine/valine dehydrogenase signature | 113 | 127 | GO:0006520|GO:0016491 |
| PRINTS | Glutamate/leucine/phenylalanine/valine dehydrogenase signature | 234 | 254 | GO:0006520|GO:0016491 |
| Pfam | Glu/Leu/Phe/Val dehydrogenase, dimerisation domain | 33 | 184 | GO:0006520|GO:0016491 |
| SMART | ELFV_dehydrog_3 | 203 | 433 | GO:0006520|GO:0016491 |
| CDD | NAD_bind_1_Glu_DH | 201 | 427 | GO:0016639 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Name | Match Start | Match End | E-value |
|---|---|---|---|---|
| Null | Null | Null | Null | Null |
Interpro hits
| Interpro Accession | Name | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR006096 | Pfam | 201 | 433 | Glutamate/phenylalanine/leucine/valine dehydrogenase, C-terminal |
| IPR036291 | SUPERFAMILY | 201 | 434 | NAD(P)-binding domain superfamily |
| IPR014362 | PIRSF | 94 | 436 | Glutamate dehydrogenase |
| IPR014362 | PIRSF | 1 | 98 | Glutamate dehydrogenase |
| IPR006095 | PRINTS | 361 | 372 | Glutamate/phenylalanine/leucine/valine dehydrogenase |
| IPR006095 | PRINTS | 192 | 214 | Glutamate/phenylalanine/leucine/valine dehydrogenase |
| IPR006095 | PRINTS | 113 | 127 | Glutamate/phenylalanine/leucine/valine dehydrogenase |
| IPR006095 | PRINTS | 234 | 254 | Glutamate/phenylalanine/leucine/valine dehydrogenase |
| IPR006097 | Pfam | 33 | 184 | Glutamate/phenylalanine/leucine/valine dehydrogenase, dimerisation domain |
| IPR006096 | SMART | 203 | 433 | Glutamate/phenylalanine/leucine/valine dehydrogenase, C-terminal |
| IPR033524 | ProSitePatterns | 121 | 134 | Leu/Phe/Val dehydrogenases active site |
| IPR033922 | CDD | 201 | 427 | NAD(P) binding domain of glutamate dehydrogenase |
Gene Structure
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| XM_028260725 | 99.902 | 1018 | 0.0 |
| XM_028260725 | 100 | 222 | 7.46e-110 |