Data Related Infomation
Camellia sinensis var. sinensis cv. Shuchazao TEA006847.1 TRI CSS Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | No Gene Product Found |
| Locus Name: | TEA006847.1 |
KEGG Orthology
K22932 K13225 K20619 K23806 K09755 K15472 K22640 K00517 K20618 K15099 K20767 K13226 K21693 K13267 K00495 K17691 K17709 K13385 K23810 K21714 K16084 K22634 K15805 K20662 K24179 K12640 K20544 K17728 K17729 K17730 K13029 K23814 K17690 K13083 K05280 K20617 K17689 K15089 K23180 K13260 K11868 K16085 K12153 K13261 K15814 K15800 K07408 K23136 K23807 K20556 K22639 K17683 K20563 K23135 K17961 K23805 K20770 K09754 K01773 K20564 K17651 K20623 K23179 K22983 K20496 K24188 K20562 K17718 K17719 K13401 K11813 K24187 K17721 K11812 K07433
Gene Family
p450
Gene Attributes
| Chromosome: | chr15 |
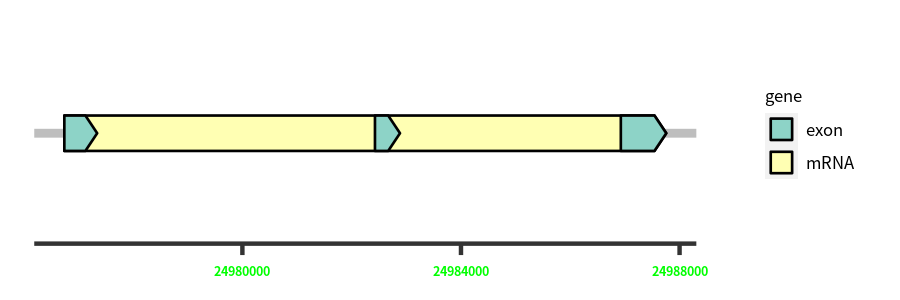
| Cds Coordinates: | 24976742-24987756 |
| Nucleotide length: | 11014 |
| Prodicted protein length: | 518 |
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| Gene3D | Cytochrome p450 | 26 | 513 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 453 | 476 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 319 | 345 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 362 | 380 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 299 | 316 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 403 | 427 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 443 | 453 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 63 | 82 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 87 | 108 | GO:0005506|GO:0016705|GO:0020037 |
| Pfam | Cytochrome P450 | 36 | 494 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | P450 superfamily signature | 363 | 374 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | P450 superfamily signature | 310 | 327 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | P450 superfamily signature | 444 | 453 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | P450 superfamily signature | 453 | 464 | GO:0005506|GO:0016705|GO:0020037 |
| ProSitePatterns | Cytochrome P450 cysteine heme-iron ligand signature. | 446 | 455 | GO:0016705 |
| SUPERFAMILY | Cytochrome P450 | 28 | 510 | GO:0005506|GO:0016705|GO:0020037 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Name | Match Start | Match End | E-value |
|---|---|---|---|---|
| Null | Null | Null | Null | Null |
Interpro hits
| Interpro Accession | Name | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR036396 | Gene3D | 26 | 513 | Cytochrome P450 superfamily |
| IPR002401 | PRINTS | 453 | 476 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 319 | 345 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 362 | 380 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 299 | 316 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 403 | 427 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 443 | 453 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 63 | 82 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 87 | 108 | Cytochrome P450, E-class, group I |
| IPR001128 | Pfam | 36 | 494 | Cytochrome P450 |
| IPR001128 | PRINTS | 363 | 374 | Cytochrome P450 |
| IPR001128 | PRINTS | 310 | 327 | Cytochrome P450 |
| IPR001128 | PRINTS | 444 | 453 | Cytochrome P450 |
| IPR001128 | PRINTS | 453 | 464 | Cytochrome P450 |
| IPR017972 | ProSitePatterns | 446 | 455 | Cytochrome P450, conserved site |
| IPR036396 | SUPERFAMILY | 28 | 510 | Cytochrome P450 superfamily |
Gene Structure
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| XM_028256484 | 100 | 1557 | 0.0 |
| KY615695 | 99.872 | 1557 | 0.0 |
| KP347675 | 99.422 | 1557 | 0.0 |
| KT180309 | 99.358 | 1557 | 0.0 |
| GQ438849 | 99.101 | 1557 | 0.0 |
| KR902747 | 98.972 | 1557 | 0.0 |
| HQ290518 | 97.495 | 1557 | 0.0 |