Data Related Infomation
Camellia sinensis var. sinensis cv. Shuchazao TEA032123.1 TRI CSS Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | No Gene Product Found |
| Locus Name: | TEA032123.1 |
KEGG Orthology
Gene Family
Gln-synt_C,Gln-synt_N
Gene Attributes
| Chromosome: | chr8 |
| Cds Coordinates: | 54825681-54831377 |
| Nucleotide length: | 5696 |
| Prodicted protein length: | 365 |
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| Pfam | Glutamine synthetase, catalytic domain | 108 | 358 | GO:0004356|GO:0006807 |
| Gene3D | Glutamine synthetase | 1 | 103 | GO:0004356|GO:0006542|GO:0006807 |
| SUPERFAMILY | Glutamine synthetase, N-terminal domain | 14 | 102 | GO:0004356|GO:0006542|GO:0006807 |
| SMART | Gln_synt_C_2 | 103 | 360 | GO:0004356|GO:0006807 |
| Pfam | Glutamine synthetase, beta-Grasp domain | 32 | 97 | GO:0004356|GO:0006542|GO:0006807 |
| SUPERFAMILY | Glutamine synthetase/guanido kinase | 103 | 362 | GO:0003824 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Name | Match Start | Match End | E-value |
|---|---|---|---|---|
| Null | Null | Null | Null | Null |
Interpro hits
| Interpro Accession | Name | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR008146 | Pfam | 108 | 358 | Glutamine synthetase, catalytic domain |
| IPR036651 | Gene3D | 1 | 103 | Glutamine synthetase, N-terminal domain superfamily |
| IPR036651 | SUPERFAMILY | 14 | 102 | Glutamine synthetase, N-terminal domain superfamily |
| IPR008146 | SMART | 103 | 360 | Glutamine synthetase, catalytic domain |
| IPR008147 | Pfam | 32 | 97 | Glutamine synthetase, beta-Grasp domain |
| IPR014746 | SUPERFAMILY | 103 | 362 | Glutamine synthetase/guanido kinase, catalytic domain |
| IPR027302 | ProSitePatterns | 55 | 72 | Glutamine synthetase, N-terminal conserved site |
Gene Structure
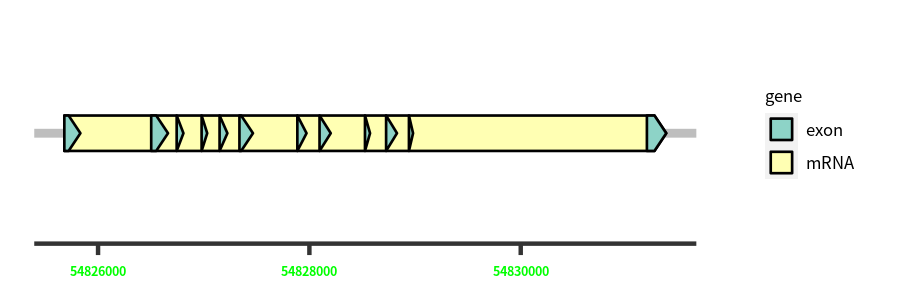
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| XM_028256209 | 100 | 720 | 0.0 |
| XM_028256209 | 100 | 353 | 0.0 |
| MG778705 | 99.583 | 720 | 0.0 |
| MG778705 | 99.433 | 353 | 2.02e-179 |
| KY649470 | 99.583 | 720 | 0.0 |
| KY649470 | 99.433 | 353 | 2.02e-179 |
| GU462131 | 99.583 | 720 | 0.0 |
| GU462131 | 99.433 | 353 | 2.02e-179 |
| AB115184 | 99.583 | 720 | 0.0 |
| AB115184 | 99.433 | 353 | 2.02e-179 |
| EU284131 | 99.444 | 720 | 0.0 |
| EU284131 | 99.433 | 353 | 2.02e-179 |
| EF055882 | 99.167 | 720 | 0.0 |
| EF055882 | 98.584 | 353 | 2.04e-174 |
| XR_003642679 | 96.842 | 190 | 3.87e-82 |
| XR_003642678 | 96.842 | 190 | 3.87e-82 |
| XR_003642677 | 96.842 | 190 | 3.87e-82 |
| XR_003642676 | 96.842 | 190 | 3.87e-82 |
| XM_028245405 | 96.842 | 190 | 3.87e-82 |
| AB597267 | 99.351 | 154 | 1.83e-70 |
| AB597267 | 98.529 | 68 | 1.17e-22 |
| AB597266 | 99.351 | 154 | 1.83e-70 |
| AB597266 | 98.529 | 68 | 1.17e-22 |
| AB597265 | 99.351 | 154 | 1.83e-70 |
| AB597265 | 98.529 | 68 | 1.17e-22 |
| AB597264 | 99.351 | 154 | 1.83e-70 |
| AB597264 | 97.059 | 68 | 5.45e-21 |
| AB597268 | 98.052 | 154 | 3.96e-67 |
| AB597268 | 98.529 | 68 | 1.17e-22 |