Data Related Infomation
Camellia sinensis var. sinensis cv. Shuchazao TEA034036.1 TRI CSS Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | Peptidase S10, serine carboxypeptidase |
| Locus Name: | TEA034036.1 |
Gene Family
Peptidase_S10
Gene Attributes
| Chromosome: | chr6 |
| Cds Coordinates: | 201178747-201187402 |
| Nucleotide length: | 8655 |
| Prodicted protein length: |
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| PRINTS | Carboxypeptidase C serine protease (S10) family signature | 147 | 157 | GO:0004185|GO:0006508 |
| PRINTS | Carboxypeptidase C serine protease (S10) family signature | 183 | 208 | GO:0004185|GO:0006508 |
| PRINTS | Carboxypeptidase C serine protease (S10) family signature | 510 | 523 | GO:0004185|GO:0006508 |
| PANTHER | SERINE PROTEASE FAMILY S10 SERINE CARBOXYPEPTIDASE | 20 | 481 | GO:0004185|GO:0006508 |
| Pfam | Serine carboxypeptidase | 46 | 481 | GO:0004185|GO:0006508 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Name | Match Start | Match End | E-value |
|---|---|---|---|---|
| PF00450.23 | 26 | 538 | Peptidase S10, serine carboxypeptidase | 1e-5 |
Interpro hits
| Interpro Accession | Name | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR029058 | Gene3D | 38 | 351 | Alpha/Beta hydrolase fold |
| IPR001563 | PRINTS | 147 | 157 | Peptidase S10, serine carboxypeptidase |
| IPR001563 | PRINTS | 183 | 208 | Peptidase S10, serine carboxypeptidase |
| IPR001563 | PRINTS | 510 | 523 | Peptidase S10, serine carboxypeptidase |
| IPR001563 | PANTHER | 20 | 481 | Peptidase S10, serine carboxypeptidase |
| IPR001563 | Pfam | 46 | 481 | Peptidase S10, serine carboxypeptidase |
| IPR029058 | SUPERFAMILY | 42 | 480 | Alpha/Beta hydrolase fold |
Gene Structure
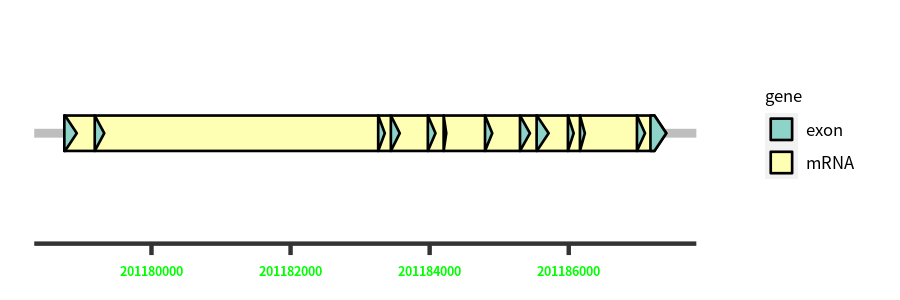
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| XM_028223563 | 100 | 245 | 1.51e-122 |
| XM_028223563 | 99.412 | 170 | 3.47e-79 |
| XM_028223563 | 100 | 144 | 2.12e-66 |
| XM_028223563 | 100 | 69 | 1.04e-24 |
| KM289196 | 100 | 245 | 1.51e-122 |
| KM289196 | 99.412 | 170 | 3.47e-79 |
| KM289196 | 100 | 144 | 2.12e-66 |
| KM289196 | 98.551 | 69 | 4.85e-23 |
| KX267893 | 99.184 | 245 | 3.28e-119 |
| KX267893 | 98.824 | 170 | 1.61e-77 |
| KX267893 | 100 | 144 | 2.12e-66 |
| KX267893 | 95.652 | 69 | 1.05e-19 |
| XM_028223557 | 97.959 | 245 | 3.30e-114 |
| XM_028223557 | 98.235 | 170 | 7.51e-76 |
| XM_028223557 | 98.611 | 144 | 4.58e-63 |
| XM_028223557 | 100 | 69 | 1.04e-24 |
| XM_028240029 | 97.959 | 245 | 3.30e-114 |
| XM_028240029 | 97.647 | 170 | 3.49e-74 |
| XM_028240029 | 98.611 | 144 | 4.58e-63 |
| XM_028240029 | 97.101 | 69 | 2.26e-21 |
| XM_028223564 | 100 | 245 | 1.51e-122 |
| XM_028223564 | 96.471 | 255 | 2.57e-110 |
| XM_028223564 | 99.412 | 170 | 3.47e-79 |
| XM_028223564 | 100 | 144 | 2.12e-66 |
| XM_028223564 | 100 | 69 | 1.04e-24 |
| XM_028240030 | 97.959 | 245 | 3.30e-114 |
| XM_028240030 | 97.647 | 170 | 3.49e-74 |
| XM_028240030 | 98.611 | 144 | 4.58e-63 |
| XM_028240030 | 97.101 | 69 | 2.26e-21 |
| XM_028223539 | 95.102 | 245 | 1.56e-102 |
| XM_028223539 | 95.266 | 169 | 5.89e-67 |
| XM_028223536 | 98.667 | 75 | 8.06e-26 |