Data Related Infomation
Camellia sinensis var. sinensis cv. Shuchazao TEA034056.1 TRI CSS Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | Peptidase S10, serine carboxypeptidase |
| Locus Name: | TEA034056.1 |
KEGG Orthology
Gene Family
Peptidase_S10
Gene Attributes
| Chromosome: | chr7 |
| Cds Coordinates: | 133432680-133456447 |
| Nucleotide length: | 23767 |
| Prodicted protein length: |
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| PRINTS | Carboxypeptidase C serine protease (S10) family signature | 222 | 247 | GO:0004185|GO:0006508 |
| PRINTS | Carboxypeptidase C serine protease (S10) family signature | 566 | 579 | GO:0004185|GO:0006508 |
| PRINTS | Carboxypeptidase C serine protease (S10) family signature | 184 | 194 | GO:0004185|GO:0006508 |
| Pfam | Serine carboxypeptidase | 339 | 654 | GO:0004185|GO:0006508 |
| Pfam | Serine carboxypeptidase | 104 | 258 | GO:0004185|GO:0006508 |
| PANTHER | SERINE PROTEASE FAMILY S10 SERINE CARBOXYPEPTIDASE | 91 | 260 | GO:0004185|GO:0006508 |
| PANTHER | SERINE PROTEASE FAMILY S10 SERINE CARBOXYPEPTIDASE | 23 | 66 | GO:0004185|GO:0006508 |
| PANTHER | SERINE PROTEASE FAMILY S10 SERINE CARBOXYPEPTIDASE | 335 | 409 | GO:0004185|GO:0006508 |
| PANTHER | SERINE PROTEASE FAMILY S10 SERINE CARBOXYPEPTIDASE | 503 | 656 | GO:0004185|GO:0006508 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Name | Match Start | Match End | E-value |
|---|---|---|---|---|
| PF00450.23 | 3 | 657 | Peptidase S10, serine carboxypeptidase | 1e-5 |
Interpro hits
| Interpro Accession | Name | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR029058 | Gene3D | 315 | 445 | Alpha/Beta hydrolase fold |
| IPR001563 | PRINTS | 222 | 247 | Peptidase S10, serine carboxypeptidase |
| IPR001563 | PRINTS | 566 | 579 | Peptidase S10, serine carboxypeptidase |
| IPR001563 | PRINTS | 184 | 194 | Peptidase S10, serine carboxypeptidase |
| IPR029058 | SUPERFAMILY | 331 | 654 | Alpha/Beta hydrolase fold |
| IPR029058 | SUPERFAMILY | 104 | 258 | Alpha/Beta hydrolase fold |
| IPR001563 | Pfam | 339 | 654 | Peptidase S10, serine carboxypeptidase |
| IPR001563 | Pfam | 104 | 258 | Peptidase S10, serine carboxypeptidase |
| IPR001563 | PANTHER | 91 | 260 | Peptidase S10, serine carboxypeptidase |
| IPR001563 | PANTHER | 23 | 66 | Peptidase S10, serine carboxypeptidase |
| IPR001563 | PANTHER | 335 | 409 | Peptidase S10, serine carboxypeptidase |
| IPR001563 | PANTHER | 503 | 656 | Peptidase S10, serine carboxypeptidase |
Gene Structure
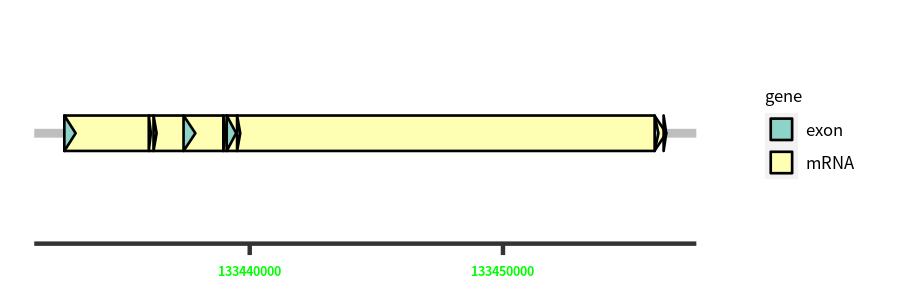
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| XM_028219599 | 95.923 | 466 | 0.0 |
| XM_028219599 | 100 | 191 | 1.94e-92 |
| XM_028219599 | 100 | 155 | 1.99e-72 |
| XM_028219599 | 100 | 84 | 5.86e-33 |
| XM_028219599 | 100 | 73 | 7.63e-27 |
| XM_028219599 | 100 | 40 | 1.69e-08 |
| XM_028219598 | 95.923 | 466 | 0.0 |
| XM_028219598 | 100 | 191 | 1.94e-92 |
| XM_028219598 | 100 | 155 | 1.99e-72 |
| XM_028219598 | 100 | 84 | 5.86e-33 |
| XM_028219598 | 100 | 73 | 7.63e-27 |
| XM_028219598 | 100 | 40 | 1.69e-08 |
| XM_028219597 | 95.923 | 466 | 0.0 |
| XM_028219597 | 100 | 191 | 1.94e-92 |
| XM_028219597 | 100 | 155 | 1.99e-72 |
| XM_028219597 | 100 | 84 | 5.86e-33 |
| XM_028219597 | 100 | 73 | 7.63e-27 |
| XM_028219597 | 100 | 40 | 1.69e-08 |
| XR_003634699 | 95.923 | 466 | 0.0 |
| XR_003634699 | 100 | 191 | 1.94e-92 |
| XR_003634699 | 100 | 155 | 1.99e-72 |
| XR_003634699 | 100 | 113 | 4.43e-49 |
| XR_003634699 | 100 | 84 | 5.86e-33 |
| XR_003634699 | 100 | 73 | 7.63e-27 |
| XR_003634699 | 100 | 43 | 3.63e-10 |
| XR_003634699 | 100 | 40 | 1.69e-08 |
| XM_028219596 | 95.923 | 466 | 0.0 |
| XM_028219596 | 100 | 191 | 1.94e-92 |
| XM_028219596 | 100 | 155 | 1.99e-72 |
| XM_028219596 | 100 | 84 | 5.86e-33 |
| XM_028219596 | 100 | 73 | 7.63e-27 |
| XM_028219596 | 100 | 40 | 1.69e-08 |
| XR_003634698 | 95.923 | 466 | 0.0 |
| XR_003634698 | 100 | 191 | 1.94e-92 |
| XR_003634698 | 100 | 155 | 1.99e-72 |
| XR_003634698 | 100 | 84 | 5.86e-33 |
| XR_003634698 | 100 | 73 | 7.63e-27 |
| XR_003634698 | 100 | 40 | 1.69e-08 |
| XR_003634697 | 95.923 | 466 | 0.0 |
| XR_003634697 | 100 | 191 | 1.94e-92 |
| XR_003634697 | 100 | 155 | 1.99e-72 |
| XR_003634697 | 100 | 84 | 5.86e-33 |
| XR_003634697 | 100 | 73 | 7.63e-27 |
| XR_003634697 | 100 | 40 | 1.69e-08 |
| XM_028219595 | 95.923 | 466 | 0.0 |
| XM_028219595 | 100 | 191 | 1.94e-92 |
| XM_028219595 | 100 | 155 | 1.99e-72 |
| XM_028219595 | 100 | 84 | 5.86e-33 |
| XM_028219595 | 100 | 73 | 7.63e-27 |
| XM_028219595 | 100 | 40 | 1.69e-08 |
| XM_028198108 | 95.708 | 466 | 0.0 |
| XM_028198108 | 100 | 191 | 1.94e-92 |
| XM_028198108 | 98.907 | 183 | 4.22e-84 |
| XM_028198108 | 100 | 40 | 1.69e-08 |
| XM_028198107 | 95.708 | 466 | 0.0 |
| XM_028198107 | 100 | 191 | 1.94e-92 |
| XM_028198107 | 100 | 155 | 1.99e-72 |
| XM_028198107 | 100 | 84 | 5.86e-33 |
| XM_028198107 | 100 | 73 | 7.63e-27 |
| XM_028198107 | 100 | 40 | 1.69e-08 |
| XM_028265729 | 100 | 155 | 1.99e-72 |
| XM_028265729 | 99.13 | 115 | 1.59e-48 |
| XM_028265729 | 100 | 84 | 5.86e-33 |
| XM_028265729 | 100 | 73 | 7.63e-27 |
| XM_028265729 | 100 | 40 | 1.69e-08 |