Data Related Infomation
Camellia sinensis var. sinensis cv. Shuchazao CSS0002737.1 AHAU CSS Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | No Gene Product Found |
| Locus Name: | CSS0002737.1 |
KEGG Orthology
Gene Family
p450
Gene Attributes
| Chromosome: | Chr4 |
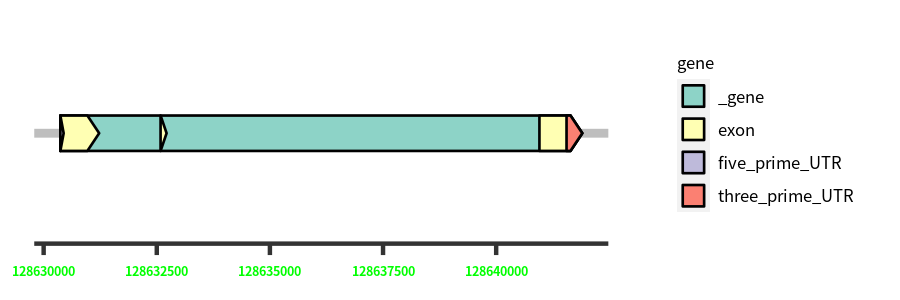
| Cds Coordinates: | 128630369-128641902 |
| Nucleotide length: | 11533 |
| Prodicted protein length: | 505 |
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| PRINTS | P450 superfamily signature | 306 | 323 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | P450 superfamily signature | 359 | 370 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | P450 superfamily signature | 438 | 447 | GO:0005506|GO:0016705|GO:0020037 |
| SUPERFAMILY | Cytochrome P450 | 34 | 503 | GO:0005506|GO:0016705|GO:0020037 |
| Gene3D | Cytochrome p450 | 24 | 501 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 181 | 199 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 315 | 341 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 399 | 423 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 62 | 81 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 447 | 470 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 295 | 312 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 86 | 107 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 358 | 376 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 437 | 447 | GO:0005506|GO:0016705|GO:0020037 |
| Pfam | Cytochrome P450 | 34 | 495 | GO:0005506|GO:0016705|GO:0020037 |
| ProSitePatterns | Cytochrome P450 cysteine heme-iron ligand signature. | 440 | 449 | GO:0016705 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Name | Match Start | Match End | E-value |
|---|---|---|---|---|
| Null | Null | Null | Null | Null |
Interpro hits
| Interpro Accession | Name | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR001128 | PRINTS | 306 | 323 | Cytochrome P450 |
| IPR001128 | PRINTS | 359 | 370 | Cytochrome P450 |
| IPR001128 | PRINTS | 438 | 447 | Cytochrome P450 |
| IPR036396 | SUPERFAMILY | 34 | 503 | Cytochrome P450 superfamily |
| IPR036396 | Gene3D | 24 | 501 | Cytochrome P450 superfamily |
| IPR002401 | PRINTS | 181 | 199 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 315 | 341 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 399 | 423 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 62 | 81 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 447 | 470 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 295 | 312 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 86 | 107 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 358 | 376 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 437 | 447 | Cytochrome P450, E-class, group I |
| IPR001128 | Pfam | 34 | 495 | Cytochrome P450 |
| IPR017972 | ProSitePatterns | 440 | 449 | Cytochrome P450, conserved site |
Gene Structure
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| KY615676 | 99.868 | 1518 | 0.0 |
| XM_028200844 | 99.539 | 1518 | 0.0 |
| KC357588 | 98.946 | 1518 | 0.0 |