Data Related Infomation
Camellia sinensis var. sinensis cv. Shuchazao CSS0030597.1 AHAU CSS Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | No Gene Product Found |
| Locus Name: | CSS0030597.1 |
KEGG Orthology
Gene Family
Chal_sti_synt_N,Chal_sti_synt_C,ACP_syn_III_C,ACP_syn_III,FAE1_CUT1_RppA
Gene Attributes
| Chromosome: | Chr5 |
| Cds Coordinates: | 148140104-148145948 |
| Nucleotide length: | 5844 |
| Prodicted protein length: | 389 |
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| Gene3D | - | 1 | 235 | GO:0016746 |
| PANTHER | HYDROXYMETHYLGLUTARYL-COA SYNTHASE | 2 | 388 | GO:0009058|GO:0016747 |
| Gene3D | - | 236 | 389 | GO:0016746 |
| SUPERFAMILY | Thiolase-like | 1 | 233 | GO:0016746 |
| PIRSF | PKS_III | 2 | 389 | GO:0009058|GO:0016747 |
| SUPERFAMILY | Thiolase-like | 235 | 386 | GO:0016746 |
| ProSitePatterns | Chalcone and stilbene synthases active site. | 156 | 172 | GO:0009058|GO:0016746 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Name | Match Start | Match End | E-value |
|---|---|---|---|---|
| Null | Null | Null | Null | Null |
Interpro hits
| Interpro Accession | Name | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR016039 | Gene3D | 1 | 235 | Thiolase-like |
| IPR011141 | PANTHER | 2 | 388 | Polyketide synthase, type III |
| IPR016039 | Gene3D | 236 | 389 | Thiolase-like |
| IPR016039 | SUPERFAMILY | 1 | 233 | Thiolase-like |
| IPR011141 | PIRSF | 2 | 389 | Polyketide synthase, type III |
| IPR001099 | Pfam | 5 | 228 | Chalcone/stilbene synthase, N-terminal |
| IPR016039 | SUPERFAMILY | 235 | 386 | Thiolase-like |
| IPR012328 | Pfam | 238 | 387 | Chalcone/stilbene synthase, C-terminal |
| IPR018088 | ProSitePatterns | 156 | 172 | Chalcone/stilbene synthase, active site |
Gene Structure
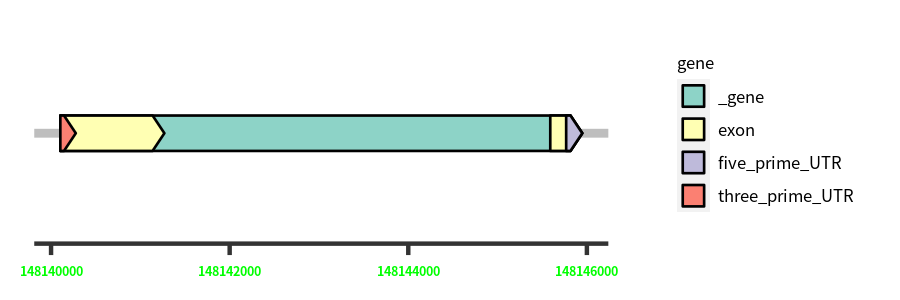
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| XM_028224715 | 99.744 | 1170 | 0.0 |
| KC357708 | 99.658 | 1170 | 0.0 |
| KY615682 | 99.573 | 1170 | 0.0 |
| D26594 | 99.573 | 1170 | 0.0 |
| MG720805 | 98.974 | 1170 | 0.0 |
| MG720807 | 98.462 | 1170 | 0.0 |
| MG720806 | 98.376 | 1170 | 0.0 |
| HQ704701 | 98.291 | 1170 | 0.0 |
| JN944574 | 98.12 | 1170 | 0.0 |
| MG720802 | 99.213 | 762 | 0.0 |
| MG720792 | 99.213 | 762 | 0.0 |
| AY146708 | 98.454 | 776 | 0.0 |
| MG720803 | 98.95 | 762 | 0.0 |
| GU722502 | 99.862 | 724 | 0.0 |
| AY146704 | 97.552 | 776 | 0.0 |
| GU722466 | 99.724 | 724 | 0.0 |
| GU722451 | 99.724 | 724 | 0.0 |
| GU722480 | 99.586 | 724 | 0.0 |
| GU722476 | 99.448 | 724 | 0.0 |
| GU722449 | 99.448 | 724 | 0.0 |
| GU722467 | 99.309 | 724 | 0.0 |
| GU722460 | 99.171 | 724 | 0.0 |
| GU722453 | 99.171 | 724 | 0.0 |
| GU722510 | 99.033 | 724 | 0.0 |
| GU722507 | 99.033 | 724 | 0.0 |
| GU722458 | 99.033 | 724 | 0.0 |
| GU722492 | 99.033 | 724 | 0.0 |
| GU722491 | 99.033 | 724 | 0.0 |
| GU722486 | 99.033 | 724 | 0.0 |
| GU722475 | 99.033 | 724 | 0.0 |
| GU722474 | 99.033 | 724 | 0.0 |
| GU722464 | 99.033 | 724 | 0.0 |
| GU722463 | 99.033 | 724 | 0.0 |
| GU722456 | 99.033 | 724 | 0.0 |
| GU722450 | 99.033 | 724 | 0.0 |
| GU722511 | 98.895 | 724 | 0.0 |
| GU722465 | 98.895 | 724 | 0.0 |
| GU722499 | 98.895 | 724 | 0.0 |
| GU722496 | 98.895 | 724 | 0.0 |
| GU722494 | 98.895 | 724 | 0.0 |
| GU722481 | 98.895 | 724 | 0.0 |
| GU722461 | 98.895 | 724 | 0.0 |
| GU722457 | 98.895 | 724 | 0.0 |
| GU722503 | 98.757 | 724 | 0.0 |
| GU722469 | 98.757 | 724 | 0.0 |
| GU722488 | 98.757 | 724 | 0.0 |
| GU722472 | 98.757 | 724 | 0.0 |
| GU722483 | 98.757 | 724 | 0.0 |
| GU722473 | 98.757 | 724 | 0.0 |
| GU722468 | 98.757 | 724 | 0.0 |
| GU722512 | 98.619 | 724 | 0.0 |
| GU722498 | 98.619 | 724 | 0.0 |
| GU722495 | 98.619 | 724 | 0.0 |
| GU722493 | 98.619 | 724 | 0.0 |
| GU722490 | 98.619 | 724 | 0.0 |
| GU722477 | 98.619 | 724 | 0.0 |
| GU722471 | 98.619 | 724 | 0.0 |
| GU722454 | 98.619 | 724 | 0.0 |
| GU722501 | 98.481 | 724 | 0.0 |
| GU722504 | 98.343 | 724 | 0.0 |
| GU722470 | 98.343 | 724 | 0.0 |
| GU722462 | 98.343 | 724 | 0.0 |
| GU722459 | 98.343 | 724 | 0.0 |
| GU722448 | 98.343 | 724 | 0.0 |
| GU722505 | 98.204 | 724 | 0.0 |
| GU722484 | 98.204 | 724 | 0.0 |
| GU722482 | 98.204 | 724 | 0.0 |
| GU722478 | 98.204 | 724 | 0.0 |
| GU722489 | 98.066 | 724 | 0.0 |
| GU722452 | 98.066 | 724 | 0.0 |
| GU722455 | 97.928 | 724 | 0.0 |
| GU722508 | 97.79 | 724 | 0.0 |
| GU722497 | 97.79 | 724 | 0.0 |
| GU722485 | 97.652 | 724 | 0.0 |
| GU722513 | 96.271 | 724 | 0.0 |
| AY570930 | 97.006 | 334 | 6.22e-155 |
| GU932717 | 97.865 | 281 | 3.85e-132 |