Data Related Infomation
Camellia sinensis var. sinensis cv. Shuchazao CSS0050330.1 AHAU CSS Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | No Gene Product Found |
| Locus Name: | CSS0050330.1 |
Gene Family
GATase_2,Glu_synthase,Glu_syn_central,GXGXG,Fer4_20,Pyr_redox_2,NAD_binding_8,Pyr_redox,FAD_oxidored,Pyr_redox_3
Gene Attributes
| Chromosome: | Chr1 |
| Cds Coordinates: | 103140344-103152859 |
| Nucleotide length: | 12515 |
| Prodicted protein length: | 2115 |
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| PIRSF | GOGAT | 41 | 1433 | GO:0005506|GO:0006537|GO:0010181|GO:0016040|GO:0050660 |
| PIRSF | GOGAT | 1539 | 2113 | GO:0005506|GO:0006537|GO:0010181|GO:0016040|GO:0050660 |
| PIRSF | GOGAT | 1430 | 1533 | GO:0005506|GO:0006537|GO:0010181|GO:0016040|GO:0050660 |
| Pfam | Pyridine nucleotide-disulphide oxidoreductase | 1884 | 2080 | GO:0016491 |
| Pfam | Glutamate synthase central domain | 594 | 861 | GO:0006807|GO:0015930 |
| Gene3D | Aldolase class I | 910 | 1350 | GO:0003824 |
| Gene3D | - | 1433 | 1523 | GO:0016491 |
| Gene3D | - | 1353 | 1432 | GO:0016491 |
| Pfam | Conserved region in glutamate synthase | 928 | 1297 | GO:0006537|GO:0015930|GO:0016638 |
| SUPERFAMILY | Alpha subunit of glutamate synthase, C-terminal domain | 1349 | 1518 | GO:0016491 |
| CDD | GltS_FMN | 930 | 1311 | GO:0006537|GO:0015930|GO:0016638 |
| TIGRFAM | GOGAT_sm_gam: glutamate synthase, NADH/NADPH, small subunit | 1606 | 2094 | GO:0006537|GO:0016639 |
| Gene3D | Aldolase class I | 582 | 909 | GO:0003824 |
| Gene3D | - | 1612 | 1748 | GO:0051536 |
| Pfam | GXGXG motif | 1434 | 1517 | GO:0016491 |
| Pfam | GXGXG motif | 1380 | 1430 | GO:0016491 |
| SUPERFAMILY | alpha-helical ferredoxin | 1605 | 1761 | GO:0051536 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Name | Match Start | Match End | E-value |
|---|---|---|---|---|
| Null | Null | Null | Null | Null |
Interpro hits
| Interpro Accession | Name | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR028261 | Pfam | 1627 | 1738 | Dihydroprymidine dehydrogenase domain II |
| IPR017932 | Pfam | 112 | 539 | Glutamine amidotransferase type 2 domain |
| IPR029055 | SUPERFAMILY | 112 | 542 | Nucleophile aminohydrolases, N-terminal |
| IPR012220 | PIRSF | 41 | 1433 | Glutamate synthase, eukaryotic |
| IPR012220 | PIRSF | 1539 | 2113 | Glutamate synthase, eukaryotic |
| IPR012220 | PIRSF | 1430 | 1533 | Glutamate synthase, eukaryotic |
| IPR023753 | Pfam | 1884 | 2080 | FAD/NAD(P)-binding domain |
| IPR006982 | Pfam | 594 | 861 | Glutamate synthase, central-N |
| IPR013785 | Gene3D | 910 | 1350 | Aldolase-type TIM barrel |
| IPR036188 | SUPERFAMILY | 1748 | 2092 | FAD/NAD(P)-binding domain superfamily |
| IPR017932 | ProSiteProfiles | 112 | 516 | Glutamine amidotransferase type 2 domain |
| IPR036485 | Gene3D | 1433 | 1523 | Glutamate synthase, alpha subunit, C-terminal domain superfamily |
| IPR036485 | Gene3D | 1353 | 1432 | Glutamate synthase, alpha subunit, C-terminal domain superfamily |
| IPR002932 | Pfam | 928 | 1297 | Glutamate synthase domain |
| IPR036485 | SUPERFAMILY | 1349 | 1518 | Glutamate synthase, alpha subunit, C-terminal domain superfamily |
| IPR029055 | Gene3D | 112 | 551 | Nucleophile aminohydrolases, N-terminal |
| IPR002932 | CDD | 930 | 1311 | Glutamate synthase domain |
| IPR006005 | TIGRFAM | 1606 | 2094 | Glutamate synthase, NADH/NADPH, small subunit 1 |
| IPR013785 | Gene3D | 582 | 909 | Aldolase-type TIM barrel |
| IPR009051 | Gene3D | 1612 | 1748 | Alpha-helical ferredoxin |
| IPR002489 | Pfam | 1434 | 1517 | Glutamate synthase, alpha subunit, C-terminal |
| IPR002489 | Pfam | 1380 | 1430 | Glutamate synthase, alpha subunit, C-terminal |
| IPR036188 | Gene3D | 1859 | 2024 | FAD/NAD(P)-binding domain superfamily |
| IPR009051 | SUPERFAMILY | 1605 | 1761 | Alpha-helical ferredoxin |
Gene Structure
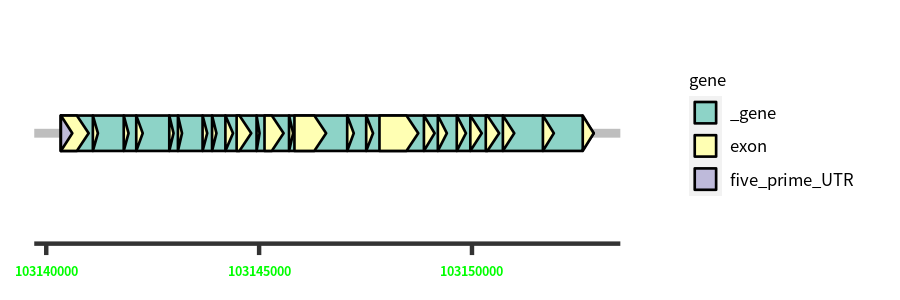
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| XM_028256854 | 98.764 | 2346 | 0.0 |
| XM_028256854 | 99.595 | 1974 | 0.0 |
| XM_028256854 | 97.747 | 1820 | 0.0 |
| XM_028256854 | 98.833 | 257 | 4.67e-123 |
| XM_028256856 | 99.595 | 1974 | 0.0 |
| XM_028256856 | 97.747 | 1820 | 0.0 |
| XM_028256856 | 98.477 | 1379 | 0.0 |
| XM_028256856 | 98.833 | 257 | 4.67e-123 |
| XM_028256855 | 99.595 | 1974 | 0.0 |
| XM_028256855 | 97.747 | 1820 | 0.0 |
| XM_028256855 | 98.477 | 1379 | 0.0 |
| XM_028256855 | 98.833 | 257 | 4.67e-123 |
| XM_028256855 | 100 | 63 | 9.00e-21 |
| XM_028254675 | 98.19 | 1050 | 0.0 |
| XM_028254674 | 98.19 | 1050 | 0.0 |
| XM_011077708 | 95.588 | 68 | 1.51e-18 |
| MH981233 | 99.118 | 567 | 0.0 |