Data Related Infomation
Camellia sinensis var. sinensis cv. Biyun GWHPAMJR005975 SCAU CSS Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | No Gene Product Found |
| Locus Name: | GWHPAMJR005975 |
KEGG Orthology
Gene Family
p450
Gene Attributes
| Chromosome: | GWHAMJR00002126 |
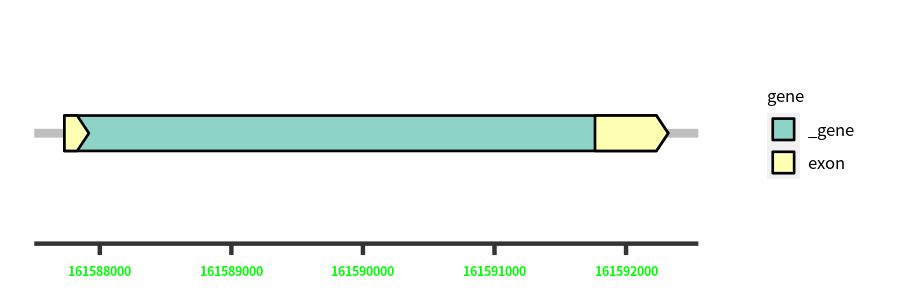
| Cds Coordinates: | 161587731-161592320 |
| Nucleotide length: | 4589 |
| Prodicted protein length: | 247 |
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| SUPERFAMILY | Cytochrome P450 | 63 | 245 | GO:0005506|GO:0016705|GO:0020037 |
| Pfam | Cytochrome P450 | 63 | 237 | GO:0005506|GO:0016705|GO:0020037 |
| Gene3D | Cytochrome p450 | 62 | 243 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | P450 superfamily signature | 66 | 79 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | P450 superfamily signature | 180 | 189 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | P450 superfamily signature | 101 | 112 | GO:0005506|GO:0016705|GO:0020037 |
| ProSitePatterns | Cytochrome P450 cysteine heme-iron ligand signature. | 182 | 191 | GO:0016705 |
| PRINTS | E-class P450 group I signature | 100 | 118 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 189 | 212 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 57 | 83 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 179 | 189 | GO:0005506|GO:0016705|GO:0020037 |
| PRINTS | E-class P450 group I signature | 141 | 165 | GO:0005506|GO:0016705|GO:0020037 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Name | Match Start | Match End | E-value |
|---|---|---|---|---|
| Null | Null | Null | Null | Null |
Interpro hits
| Interpro Accession | Name | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR036396 | SUPERFAMILY | 63 | 245 | Cytochrome P450 superfamily |
| IPR001128 | Pfam | 63 | 237 | Cytochrome P450 |
| IPR036396 | Gene3D | 62 | 243 | Cytochrome P450 superfamily |
| IPR001128 | PRINTS | 66 | 79 | Cytochrome P450 |
| IPR001128 | PRINTS | 180 | 189 | Cytochrome P450 |
| IPR001128 | PRINTS | 101 | 112 | Cytochrome P450 |
| IPR017972 | ProSitePatterns | 182 | 191 | Cytochrome P450, conserved site |
| IPR002401 | PRINTS | 100 | 118 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 189 | 212 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 57 | 83 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 179 | 189 | Cytochrome P450, E-class, group I |
| IPR002401 | PRINTS | 141 | 165 | Cytochrome P450, E-class, group I |
Gene Structure
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| XM_028200844 | 99.641 | 557 | 0.0 |
| KY615676 | 99.461 | 557 | 0.0 |
| KC357588 | 99.282 | 557 | 0.0 |
| XM_028199676 | 97.087 | 103 | 5.92e-39 |
| XM_028266404 | 95.181 | 83 | 3.62e-26 |
| XM_028263329 | 97.183 | 71 | 7.83e-23 |
| XM_028211474 | 98.214 | 56 | 3.67e-16 |
| XM_028211473 | 98.214 | 56 | 3.67e-16 |