Data Related Infomation
Camellia sinensis var. sinensis cv. Biyun GWHPAMJR009265 SCAU CSS Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | No Gene Product Found |
| Locus Name: | GWHPAMJR009265 |
KEGG Orthology
Gene Family
Methyltransf_7
Gene Attributes
| Chromosome: | GWHAMJR00000425 |
| Cds Coordinates: | 106003348-106017886 |
| Nucleotide length: | 14538 |
| Prodicted protein length: | 800 |
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| Pfam | SAM dependent carboxyl methyltransferase | 382 | 581 | GO:0008168 |
| PANTHER | S-ADENOSYL-L-METHIONINE:CARBOXYL METHYLTRANSFERASE FAMILY PROTEIN | 357 | 581 | GO:0008168 |
| PANTHER | S-ADENOSYL-L-METHIONINE:CARBOXYL METHYLTRANSFERASE FAMILY PROTEIN | 584 | 633 | GO:0008168 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Name | Match Start | Match End | E-value |
|---|---|---|---|---|
| Null | Null | Null | Null | Null |
Interpro hits
| Interpro Accession | Name | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR029063 | SUPERFAMILY | 358 | 635 | S-adenosyl-L-methionine-dependent methyltransferase |
| IPR042086 | Gene3D | 490 | 581 | Methyltransferase, alpha-helical capping domain |
| IPR005299 | Pfam | 382 | 581 | SAM dependent carboxyl methyltransferase |
| IPR005299 | PANTHER | 357 | 581 | SAM dependent carboxyl methyltransferase |
| IPR042086 | Gene3D | 582 | 633 | Methyltransferase, alpha-helical capping domain |
| IPR005299 | PANTHER | 584 | 633 | SAM dependent carboxyl methyltransferase |
Gene Structure
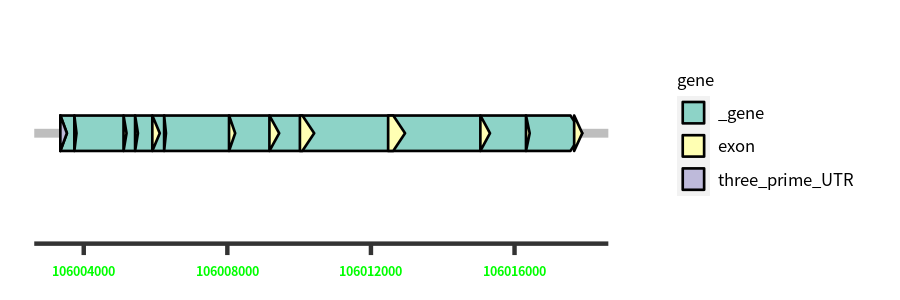
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| AB207818 | 98.37 | 675 | 0.0 |
| AB207818 | 96.667 | 180 | 3.12e-76 |
| AB207816 | 98.37 | 675 | 0.0 |
| AB207816 | 98.333 | 180 | 3.10e-81 |
| AB031281 | 98.217 | 673 | 0.0 |
| AB031281 | 97.222 | 180 | 6.71e-78 |
| XM_028205740 | 97.654 | 682 | 0.0 |
| AY907710 | 97.92 | 673 | 0.0 |
| AY907710 | 96.667 | 180 | 3.12e-76 |
| XM_028205684 | 97.768 | 672 | 0.0 |
| XM_028205684 | 97.778 | 180 | 1.44e-79 |
| XM_028205701 | 97.47 | 672 | 0.0 |
| XM_028205701 | 96.111 | 180 | 1.45e-74 |
| AB056108 | 96.731 | 673 | 0.0 |
| AB056108 | 95.48 | 177 | 3.15e-71 |
| AB207817 | 96.582 | 673 | 0.0 |
| AB297451 | 96.434 | 673 | 0.0 |
| XM_028226211 | 96.296 | 675 | 0.0 |
| XM_028226210 | 96.285 | 673 | 0.0 |
| XM_028226210 | 97.17 | 212 | 5.08e-94 |
| XM_028226210 | 96.667 | 60 | 1.22e-15 |
| XM_028226209 | 96.285 | 673 | 0.0 |
| XM_028226209 | 99.435 | 177 | 6.67e-83 |
| AB362884 | 96.137 | 673 | 0.0 |
| MN163830 | 95.976 | 671 | 0.0 |
| MN163830 | 96.045 | 177 | 6.76e-73 |
| AB362882 | 95.84 | 673 | 0.0 |
| XM_028228398 | 95.833 | 672 | 0.0 |
| XM_028228398 | 97.143 | 175 | 4.04e-75 |
| KT215399 | 95.691 | 673 | 0.0 |
| KT215399 | 96.089 | 179 | 5.23e-74 |
| KT215398 | 95.691 | 673 | 0.0 |
| KT215398 | 96.648 | 179 | 1.12e-75 |
| KT215397 | 95.691 | 673 | 0.0 |
| KT215397 | 95.531 | 179 | 2.43e-72 |
| AB362885 | 95.562 | 676 | 0.0 |
| AB362883 | 95.562 | 676 | 0.0 |
| AB362883 | 95.556 | 180 | 6.76e-73 |
| MN163831 | 95.529 | 671 | 0.0 |
| MN163831 | 97.207 | 179 | 2.42e-77 |
| AB362886 | 95.562 | 676 | 0.0 |
| XM_028209955 | 95.394 | 673 | 0.0 |
| AB031280 | 95.536 | 672 | 0.0 |
| AB031280 | 96.571 | 175 | 1.88e-73 |
| MN163829 | 96 | 175 | 8.75e-72 |
| XM_028211977 | 97.368 | 494 | 0.0 |
| JX647691 | 97.75 | 400 | 0.0 |
| JX647691 | 98.909 | 275 | 4.80e-134 |
| JX647691 | 97.222 | 180 | 6.71e-78 |
| JX647695 | 96.25 | 400 | 0.0 |
| JX647695 | 97.818 | 275 | 4.84e-129 |
| JX647695 | 99.435 | 177 | 6.67e-83 |
| JX647693 | 95.285 | 403 | 2.10e-177 |
| JX647693 | 95.29 | 276 | 2.28e-117 |
| FJ554589 | 97.067 | 375 | 2.71e-176 |
| JX647694 | 95.556 | 180 | 6.76e-73 |
| EF526217 | 97.455 | 275 | 2.25e-127 |
| EF526217 | 97.143 | 175 | 4.04e-75 |
| JX647690 | 97.455 | 275 | 2.25e-127 |
| JX647690 | 97.143 | 175 | 4.04e-75 |
| XM_028268989 | 97.578 | 289 | 3.71e-135 |
| XM_028226196 | 97.156 | 211 | 1.83e-93 |
| HM003356 | 97.222 | 180 | 6.71e-78 |
| HM003356 | 97.701 | 87 | 3.33e-31 |
| XM_028247252 | 96.648 | 179 | 1.12e-75 |
| XM_028247252 | 96.667 | 150 | 3.19e-61 |
| XM_028268333 | 95.902 | 122 | 1.17e-45 |
| XM_028268338 | 95.868 | 121 | 4.22e-45 |
| XM_028268336 | 95.868 | 121 | 4.22e-45 |
| XM_028268335 | 95.868 | 121 | 4.22e-45 |
| XM_028268334 | 98.058 | 103 | 4.25e-40 |
| XM_028268332 | 98.058 | 103 | 4.25e-40 |
| XM_028268331 | 98.058 | 103 | 4.25e-40 |