Data Related Infomation
Camellia Sinesis var. sinensis cv. Huangdan GWHPAZTZ033151 FAFU release 1 annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | No Gene Product Found |
| Locus Name: | GWHPAZTZ033151 |
KEGG Orthology
Gene Family
Epimerase
Gene Attributes
| Chromosome: | GWHAZTZ00000009 |
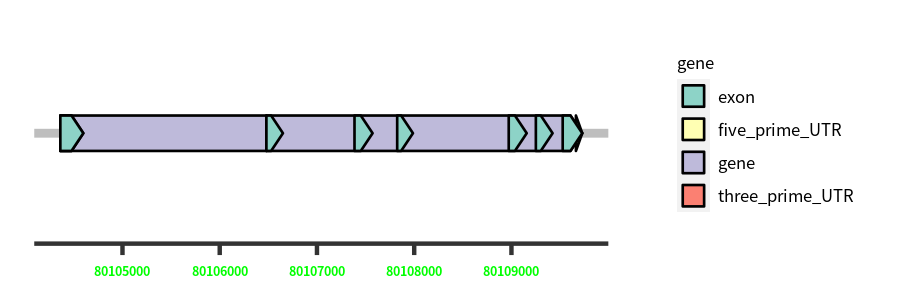
| Cds Coordinates: | 80109662-80109527 |
| Nucleotide length: | 990 |
| Prodicted protein length: | 329 |
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| Pfam | NAD dependent epimerase/dehydratase family | 14 | 254 | GO:0003824 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Name | Match Start | Match End | E-value |
|---|---|---|---|---|
| PF01370.22 | 12 | 328 | NAD dependent epimerase/dehydratase family | 1e-5 |
| PF01370.22 | 12 | 328 | NAD dependent epimerase/dehydratase family | 1e-5 |
| PF01073.20 | 12 | 328 | 3-beta hydroxysteroid dehydrogenase/isomerase family | 1e-5 |
| PF01073.20 | 12 | 328 | 3-beta hydroxysteroid dehydrogenase/isomerase family | 1e-5 |
| PF16363.6 | 12 | 328 | GDP-mannose 4,6 dehydratase | 1e-5 |
| PF16363.6 | 12 | 328 | GDP-mannose 4,6 dehydratase | 1e-5 |
| PF13460.7 | 12 | 328 | NAD(P)H-binding | 1e-5 |
| PF13460.7 | 12 | 328 | NAD(P)H-binding | 1e-5 |
| PF07993.13 | 12 | 328 | Male sterility protein | 1e-5 |
| PF07993.13 | 12 | 328 | Male sterility protein | 1e-5 |
| PF05368.14 | 12 | 328 | NmrA-like family | 1e-5 |
| PF05368.14 | 12 | 328 | NmrA-like family | 1e-5 |
| PF02719.16 | 12 | 328 | Polysaccharide biosynthesis protein | 1e-5 |
| PF02719.16 | 12 | 328 | Polysaccharide biosynthesis protein | 1e-5 |
| PF08659.11 | 12 | 328 | KR domain | 1e-5 |
| PF08659.11 | 12 | 328 | KR domain | 1e-5 |
Interpro hits
| Interpro Accession | Name | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR036291 | SUPERFAMILY | 10 | 322 | NAD(P)-binding domain superfamily |
| IPR001509 | Pfam | 14 | 254 | NAD-dependent epimerase/dehydratase |
Gene Structure
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| XM_028256593.1 | 99.192 | 990 | 0.0 |