Data Related Infomation
Camellia sinensis var. sinensis cv. Longjing43 GWHPACFB026040 TRI CSS Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | No Gene Product Found |
| Locus Name: | GWHPACFB026040 |
KEGG Orthology
Gene Family
Chal_sti_synt_N,Chal_sti_synt_C,ACP_syn_III_C,ACP_syn_III,FAE1_CUT1_RppA
Gene Attributes
| Chromosome: | GWHACFB00000009 |
| Cds Coordinates: | 120821185-120833765 |
| Nucleotide length: | 12580 |
| Prodicted protein length: | 463 |
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| Null | Null | Null | Null | Null |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Name | Match Start | Match End | E-value |
|---|---|---|---|---|
| Null | Null | Null | Null | Null |
Interpro hits
| Interpro Accession | Name | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR016039 | Gene3D | 1 | 235 | Thiolase-like |
| IPR001099 | Pfam | 5 | 228 | Chalcone/stilbene synthase, N-terminal |
| IPR016039 | Gene3D | 236 | 390 | Thiolase-like |
| IPR011141 | PANTHER | 2 | 383 | Polyketide synthase, type III |
| IPR012328 | Pfam | 238 | 384 | Chalcone/stilbene synthase, C-terminal |
| IPR016039 | SUPERFAMILY | 1 | 232 | Thiolase-like |
| IPR018088 | ProSitePatterns | 156 | 172 | Chalcone/stilbene synthase, active site |
| IPR016039 | SUPERFAMILY | 235 | 385 | Thiolase-like |
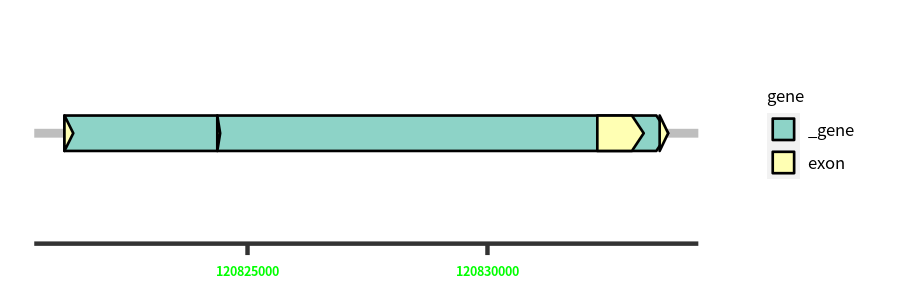
Gene Structure
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| XM_028211683 | 99.913 | 1146 | 0.0 |
| KY615681 | 99.651 | 1146 | 0.0 |
| D26593 | 99.651 | 1146 | 0.0 |
| KC357707 | 99.564 | 1146 | 0.0 |
| JQ247184 | 99.302 | 1146 | 0.0 |
| AY169403 | 99.215 | 1146 | 0.0 |
| AB512766 | 98.691 | 1146 | 0.0 |
| JN944573 | 98.429 | 1146 | 0.0 |
| HQ269804 | 98.342 | 1146 | 0.0 |
| MG720809 | 97.644 | 1146 | 0.0 |
| MG720797 | 97.644 | 1146 | 0.0 |
| MG720810 | 97.382 | 1146 | 0.0 |
| MG720798 | 97.295 | 1146 | 0.0 |
| MG720808 | 97.208 | 1146 | 0.0 |
| MG720811 | 97.12 | 1146 | 0.0 |
| MG720799 | 97.033 | 1146 | 0.0 |
| MG720800 | 96.859 | 1146 | 0.0 |
| MG720801 | 96.684 | 1146 | 0.0 |
| KC357705 | 99.69 | 968 | 0.0 |
| KC357705 | 100 | 180 | 1.77e-86 |
| JQ269640 | 98.888 | 809 | 0.0 |
| AY146698 | 97.812 | 777 | 0.0 |
| AY146703 | 97.683 | 777 | 0.0 |
| MG720804 | 98.103 | 738 | 0.0 |
| AY146706 | 96.521 | 776 | 0.0 |
| MG720793 | 97.967 | 738 | 0.0 |
| MG720795 | 97.832 | 738 | 0.0 |
| MG720796 | 97.696 | 738 | 0.0 |
| AY570926 | 99.012 | 405 | 0.0 |
| KJ866952 | 99.123 | 342 | 1.57e-171 |