Data Related Infomation
Camellia sinensis var. sinensis cv. Tieguanyin GWHPASIV022806 FAFU Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | Protein kinase domain |
| Locus Name: | GWHPASIV022806 |
KEGG Orthology
K13416 K13430 K13415 K13418 K04730 K00924 K13420 K13417 K20718 K04733 K13429 K16224 K13436 K18873 K04427 K13435 K08844 K13428 K04732 K08884 K04447 K14500 K04367 K08846 K05111 K05704 K08847 K04365 K04419 K08892 K08842 K02861 K08794 K02831 K05703 K11217 K06641 K08889 K13437 K08850 K05743 K20607 K08269 K08841 K04366 K08838 K08895 K07198 K13303 K04362 K05108 K05705 K08282 K02216 K16289 K05103 K21358 K08852 K04423 K04345 K11481 K06633 K08848 K05728 K04417 K05121 K05126 K11479 K08893 K13412 K08813 K04368 K02202 K20604 K11226 K11916 K08294 K05105 K04411 K06272 K08843 K05104 K21357 K08845 K07363 K08865 K08857 K02206 K20717 K08829 K04420 K05106 K02087 K08863 K13596 K08896 K20603 K05102 K08252 K07364 K05110 K05099 K13302 K08817 K05129 K20606 K11218 K20716 K06655 K12324 K07298 K17511 K08856 K05093 K19800 K08894 K17512 K16195 K03114 K04422 K08849 K20872 K07359 K04360 K04688 K11480 K08798 K04410 K06276 K13419 K12776 K06683 K08835 K06668 K08824 K20879 K16288 K05855 K04467 K05854 K05122 K12761 K05125 K20876 K05107 K22600 K05112 K18670 K12323 K08859 K08861 K08828 K04421 K04373 K07209 K13413 K17534 K05109 K08811 K20605 K03083 K04418 K04670 K04463 K13872 K04412 K06631 K02089 K19477 K23453 K04430 K18529 K05869 K16196 K08818 K16510 K12771 K04563 K04672 K06070 K11230 K02449 K11219 K05115 K16310 K08864 K07376 K11912 K00910 K08799 K05127 K04445 K08885 K16194 K17545 K08793 K08796 K05101 K08888 K05744 K20878 K07370 K04675 K04527 K20602 K05087 K05113 K00908 K04674 K06660 K08860 K08016 K08862 K04416 K04456 K08806 K13976 K20600 K02090 K18669 K14758 K00871 K05094 K14502 K08805 K15048 K23691 K05119 K04673 K20359 K08822 K13578 K17510 K05871 K08819 K07527 K19603 K04409 K05123 K04426 K12322 K12766 K20877 K04369 K06632 K04468 K05733 K08291 K08802 K02091 K04515 K08867 K18952 K08886 K00916 K19596 K14512 K04372 K04432 K07360 K05124 K17544 K05100 K01769 K04433 K08795 K08809 K05095 K02218 K04702 K20536 K11889 K04440 K05117 K08807 K05128 K02211 K08834 K22009 K18052 K05725 K20875 K08827 K17302 K08810 K19662 K05085 K20535 K08800 K16308 K08808 K11228 K05410 K20880 K08957 K08804 K05734 K18051 K00909 K20537 K15562 K14498 K08821 K19663 K18050 K17547 K18679 K15595 K08830 K04431 K08840 K05083 K03097 K06667 K12378 K08826 K11229 K05091 K11715 K20873 K17530 K19008 K05736 K04428 K19009 K16311 K13975 K05096 K05735 K02677 K17480 K08959 K05097 K06855 K15486 K08293 K23561 K04388 K08831 K05086 K05098 K08797 K08803 K06069 K19763 K05090 K17388 K05116 K05088 K02208 K08790 K17541 K04464 K08853 K08877 K04406 K11227 K04408 K16316 K12321 K13568 K08960 K08899 K14958 K15596 K05033 K13567 K05092 K17546 K08271 K06103 K08858 K15594 K04414 K02214 K04425 K20538 K08832 K17751 K20715 K08855 K13594 K08801 K04443 K15409 K08866 K05089 K21157 K08825 K08836 K20714 K18680 K12765 K19439 K23785 K08823 K16309 K04363 K08898 K17533 K08833 K04514 K08854 K08958 K04444 K17540 K20839 K06684 K04413 K21158 K04407 K17531 K08788 K12767 K06686 K05688 K17532 K01165 K04442 K01412 K16313 K19847 K08601 K16279 K02178 K16312 K08816 K08812 K16240 K17543 K08789 K16307 K19690
Gene Family
Pkinase
Gene Attributes
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| Pfam | Protein kinase domain | 497 | 762 | GO:0004672|GO:0005524|GO:0006468 |
| ProSiteProfiles | Protein kinase domain | 494 | 768 | GO:0004672|GO:0005524|GO:0006468 |
| SMART | Protein kinase domain | 494 | 763 | GO:0004672|GO:0005524|GO:0006468 |
| ProSitePatterns | Protein kinase, ATP binding site | 500 | 523 | GO:0005524 |
| PIRSF | S-receptor-like serine/threonine-protein kinase | 4 | 820 | GO:0004674 |
| Pfam | S-locus glycoprotein domain | 213 | 320 | GO:0048544 |
| ProSitePatterns | Serine/threonine-protein kinase, active site | 614 | 626 | GO:0004672|GO:0006468 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Annotation | Match Start | Match End | E-value |
|---|---|---|---|---|
| PF00069 | Protein kinase domain | 497 | 762 | 1e-5 |
| PF01453 | D-mannose binding lectin | 77 | 181 | 1e-5 |
| PF00954 | S-locus glycoprotein domain | 213 | 320 | 1e-5 |
| PF08276 | PAN-like domain | 352 | 409 | 1e-5 |
Interpro hits
| Interpro Accession | Database | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR000719 | Pfam | 497 | 762 | Protein kinase domain |
| IPR000719 | ProSiteProfiles | 494 | 768 | Protein kinase domain profile. |
| IPR000719 | SMART | 494 | 763 | serkin_6 |
| IPR003609 | SMART | 352 | 427 | ntp_6 |
| IPR003609 | ProSiteProfiles | 344 | 428 | PAN/Apple domain profile. |
| - | CDD | 339 | 428 | PAN_AP_plant |
| IPR001480 | ProSiteProfiles | 29 | 151 | Bulb-type lectin domain profile. |
| - | PANTHER | 9 | 794 | G-TYPE LECTIN S-RECEPTOR-LIKE SERINE/THREONINE-KINASE PLANT |
| - | Gene3D | 569 | 772 | Transferase(Phosphotransferase) domain 1 |
| IPR011009 | SUPERFAMILY | 481 | 800 | Protein kinase-like (PK-like) |
| IPR017441 | ProSitePatterns | 500 | 523 | Protein kinases ATP-binding region signature. |
| IPR024171 | PIRSF | 4 | 820 | SRK |
| IPR001480 | Pfam | 77 | 181 | D-mannose binding lectin |
| IPR001480 | SMART | 35 | 155 | blect_4 |
| - | Gene3D | 458 | 568 | Phosphorylase Kinase; domain 1 |
| - | MobiDBLite | 795 | 820 | consensus disorder prediction |
| IPR036426 | SUPERFAMILY | 63 | 219 | alpha-D-mannose-specific plant lectins |
| - | CDD | 500 | 766 | STKc_IRAK |
| IPR001480 | CDD | 35 | 155 | B_lectin |
| IPR000858 | Pfam | 213 | 320 | S-locus glycoprotein domain |
| IPR003609 | Pfam | 352 | 409 | PAN-like domain |
| IPR036426 | Gene3D | 29 | 153 | Agglutinin |
| IPR008271 | ProSitePatterns | 614 | 626 | Serine/Threonine protein kinases active-site signature. |
| - | PANTHER | 9 | 794 | S-LOCUS LECTIN KINASE FAMILY PROTEIN |
Gene Structure
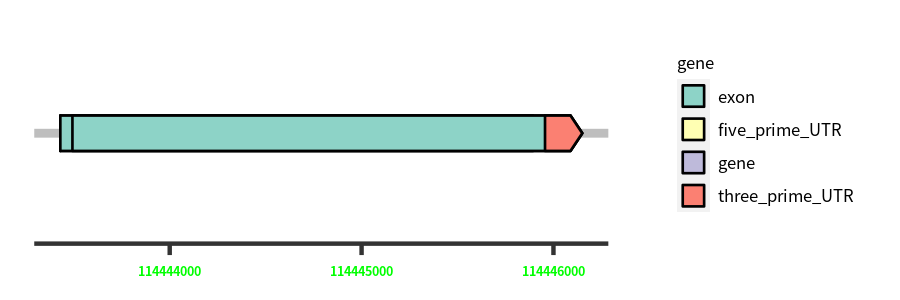
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| Null | Null | Null | Null |