Data Related Infomation
Camellia sinensis var. sinensis cv. Tieguanyin GWHPASIV025272 FAFU Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | NAD-dependent epimerase/dehydratase |
| Locus Name: | GWHPASIV025272 |
KEGG Orthology
K21102 K13082 K13265 K09753 K22098 K17741 K00091 K15891 K22320 K01784 K19997 K07748 K12408 K00070 K15856 K16045 K15894 K23557 K22025 K23558 K02473 K22252 K01710 K19267 K21332 K07118 K08679 K08678 K10046 K12454 K02377 K01709 K21271 K19660 K13313 K05901 K12453 K12448 K21211 K12449 K21793 K19073 K17716 K03274 K01711 K07071 K21214 K12441 K19068 K00067 K12455 K24312 K04791 K05281 K07535 K16652 K00065 K00046 K00300 K07124 K03793 K15789 K01043 K21883 K24002 K05829 K18009 K19652 K00023 K06988 K00034 K10620 K00293 K03366
Gene Family
Epimerase
Gene Attributes
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| Pfam | NAD-dependent epimerase/dehydratase | 12 | 201 | GO:0003824 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Annotation | Match Start | Match End | E-value |
|---|---|---|---|---|
| PF01370 | NAD dependent epimerase/dehydratase family | 12 | 201 | 1e-5 |
Interpro hits
| Interpro Accession | Database | Match Start | Match End | Annotation |
|---|---|---|---|---|
| - | PANTHER | 227 | 310 | NAD DEPENDENT EPIMERASE/DEHYDRATASE |
| - | Gene3D | 2 | 312 | - |
| - | PANTHER | 227 | 310 | ANTHOCYANIDIN REDUCTASE |
| - | CDD | 12 | 287 | FR_SDR_e |
| - | PANTHER | 10 | 229 | NAD DEPENDENT EPIMERASE/DEHYDRATASE |
| - | PANTHER | 10 | 229 | ANTHOCYANIDIN REDUCTASE |
| IPR036291 | SUPERFAMILY | 8 | 305 | NAD(P)-binding Rossmann-fold domains |
| IPR001509 | Pfam | 12 | 201 | NAD dependent epimerase/dehydratase family |
Gene Structure
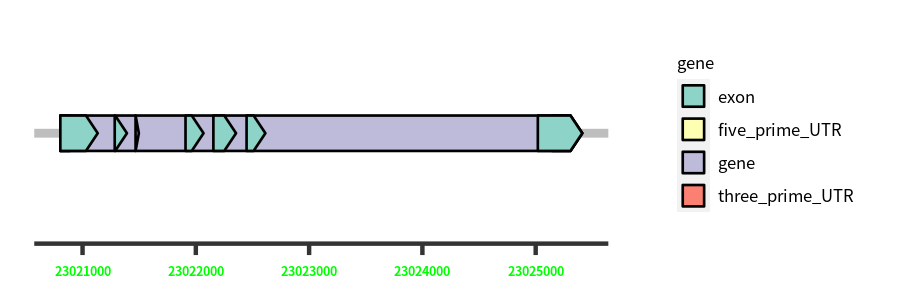
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| Null | Null | Null | Null |