Data Related Infomation
Camellia sinensis var. sinensis cv. Tieguanyin GWHPASIV034383 FAFU Release 1 Annotation
DNA Coding Sequence & Protein Sequence
Gene Identification
| Gene Product Name: | Leucine-rich repeat |
| Locus Name: | GWHPASIV034383 |
KEGG Orthology
K13466 K13420 K13465 K13468 K13471 K13416 K13470 K13446 K13428 K13437 K08844 K12796 K16340 K08399 K19613 K17256 K13459 K04306 K16224 K08462 K24055 K10159 K13415 K13457 K06839 K20718 K04308 K06838 K06555 K18809 K18808 K18807 K14142 K13418 K20599 K04309 K23533 K08123 K05401 K08119 K13023 K20359 K06260 K04307 K08120 K08460 K06850 K08118 K17550 K13417 K10130 K16362 K24500 K10160 K08843 K24492 K10348 K16351 K16665 K04247 K08461 K22038 K20230 K06261 K10641 K16358 K10169 K13460 K20237 K10168 K08121 K00924 K08130 K16360 K17568 K16661 K08127 K15069 K22529 K04985 K08128 K24095 K24493 K08124 K16666 K16355 K16606 K16354 K08125 K16356 K16659 K04249 K10171 K14050 K16357 K19750 K18647 K10270 K16660 K23946 K22449 K12805 K20493 K24491 K14319 K16717 K03021 K10411 K14485 K16226 K18648 K14284 K17579 K17567 K13453
Gene Family
LRR_8
Gene Attributes
Gene Ontology Classification
| Database | Annotation | Match Start | Match End | GO |
|---|---|---|---|---|
| Pfam | Leucine-rich repeat | 413 | 473 | GO:0005515 |
| Pfam | Leucine-rich repeat | 270 | 328 | GO:0005515 |
| Pfam | Leucine-rich repeat | 539 | 599 | GO:0005515 |
| Pfam | Leucine-rich repeat | 246 | 268 | GO:0005515 |
| Pfam | Leucine-rich repeat | 950 | 966 | GO:0005515 |
| Pfam | Leucine-rich repeat | 788 | 808 | GO:0005515 |
| Pfam | Leucine-rich repeat | 860 | 882 | GO:0005515 |
Signal Peptide
No Signal Peptide Found
PFAM hits
| Accession | Annotation | Match Start | Match End | E-value |
|---|---|---|---|---|
| PF08263 | Leucine rich repeat N-terminal domain | 34 | 72 | 1e-5 |
| PF13855 | Leucine rich repeat | 413 | 473 | 1e-5 |
| PF13855 | Leucine rich repeat | 270 | 328 | 1e-5 |
| PF13855 | Leucine rich repeat | 539 | 599 | 1e-5 |
| PF00560 | Leucine Rich Repeat | 246 | 268 | 1e-5 |
| PF00560 | Leucine Rich Repeat | 950 | 966 | 1e-5 |
| PF00560 | Leucine Rich Repeat | 788 | 808 | 1e-5 |
| PF00560 | Leucine Rich Repeat | 860 | 882 | 1e-5 |
Interpro hits
| Interpro Accession | Database | Match Start | Match End | Annotation |
|---|---|---|---|---|
| IPR003591 | SMART | 460 | 483 | LRR_typ_2 |
| IPR003591 | SMART | 244 | 267 | LRR_typ_2 |
| IPR003591 | SMART | 785 | 808 | LRR_typ_2 |
| IPR003591 | SMART | 947 | 971 | LRR_typ_2 |
| IPR003591 | SMART | 537 | 561 | LRR_typ_2 |
| IPR003591 | SMART | 113 | 138 | LRR_typ_2 |
| IPR003591 | SMART | 219 | 242 | LRR_typ_2 |
| IPR003591 | SMART | 388 | 412 | LRR_typ_2 |
| IPR003591 | SMART | 995 | 1018 | LRR_typ_2 |
| IPR003591 | SMART | 340 | 364 | LRR_typ_2 |
| IPR003591 | SMART | 292 | 316 | LRR_typ_2 |
| IPR003591 | SMART | 858 | 882 | LRR_typ_2 |
| - | SMART | 513 | 534 | LRR_sd22_2 |
| - | SMART | 292 | 311 | LRR_sd22_2 |
| - | SMART | 436 | 465 | LRR_sd22_2 |
| - | SMART | 537 | 563 | LRR_sd22_2 |
| - | SMART | 586 | 605 | LRR_sd22_2 |
| - | SMART | 858 | 884 | LRR_sd22_2 |
| - | SUPERFAMILY | 357 | 676 | RNI-like |
| - | PANTHER | 539 | 1100 | LRR RECEPTOR-LIKE SERINE/THREONINE-PROTEIN KINASE FLS2-RELATED |
| - | PANTHER | 383 | 553 | LRR RECEPTOR-LIKE SERINE/THREONINE-PROTEIN KINASE FLS2-RELATED |
| IPR032675 | Gene3D | 581 | 1043 | Ribonuclease Inhibitor |
| IPR032675 | Gene3D | 514 | 580 | Ribonuclease Inhibitor |
| - | PANTHER | 339 | 383 | LRR RECEPTOR-LIKE SERINE/THREONINE-PROTEIN KINASE FLS2-RELATED |
| - | PANTHER | 31 | 333 | LRR RECEPTOR-LIKE SERINE/THREONINE-PROTEIN KINASE FLS2 |
| - | SUPERFAMILY | 699 | 1033 | L domain-like |
| - | SUPERFAMILY | 105 | 375 | RNI-like |
| IPR013210 | Pfam | 34 | 72 | Leucine rich repeat N-terminal domain |
| - | PANTHER | 339 | 383 | LRR RECEPTOR-LIKE SERINE/THREONINE-PROTEIN KINASE FLS2 |
| - | PANTHER | 539 | 1100 | LRR RECEPTOR-LIKE SERINE/THREONINE-PROTEIN KINASE FLS2 |
| IPR032675 | Gene3D | 211 | 308 | Ribonuclease Inhibitor |
| IPR032675 | Gene3D | 27 | 210 | Ribonuclease Inhibitor |
| IPR032675 | Gene3D | 309 | 405 | Ribonuclease Inhibitor |
| - | PRINTS | 295 | 308 | Leucine-rich repeat signature |
| - | PRINTS | 537 | 550 | Leucine-rich repeat signature |
| - | PANTHER | 31 | 333 | LRR RECEPTOR-LIKE SERINE/THREONINE-PROTEIN KINASE FLS2-RELATED |
| IPR032675 | Gene3D | 406 | 510 | Ribonuclease Inhibitor |
| IPR001611 | Pfam | 413 | 473 | Leucine rich repeat |
| IPR001611 | Pfam | 270 | 328 | Leucine rich repeat |
| IPR001611 | Pfam | 539 | 599 | Leucine rich repeat |
| IPR001611 | Pfam | 246 | 268 | Leucine Rich Repeat |
| IPR001611 | Pfam | 950 | 966 | Leucine Rich Repeat |
| IPR001611 | Pfam | 788 | 808 | Leucine Rich Repeat |
| IPR001611 | Pfam | 860 | 882 | Leucine Rich Repeat |
| - | PANTHER | 383 | 553 | LRR RECEPTOR-LIKE SERINE/THREONINE-PROTEIN KINASE FLS2 |
Gene Structure
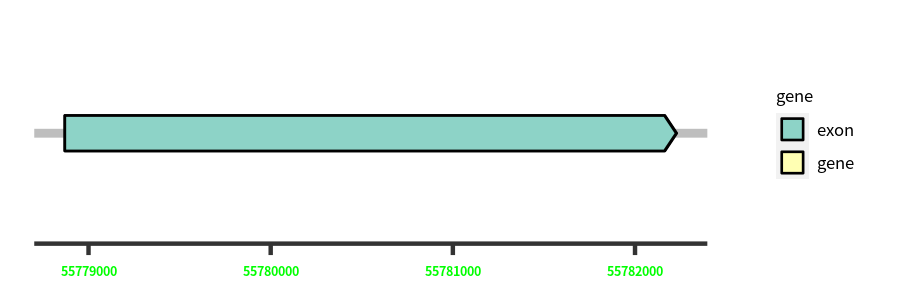
Blastn Searches(aligned nr database from NCBI)
| Accession(nr database) | %Identity | Length | E-value |
|---|---|---|---|
| Null | Null | Null | Null |